2026
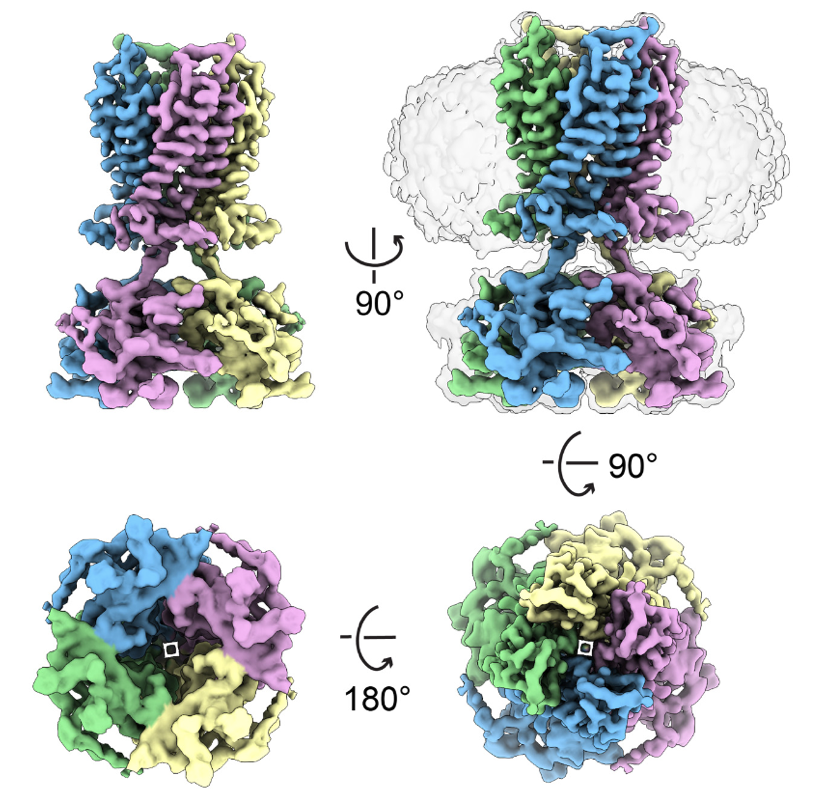
Niu Q*, Vu S*, Xu Y, Qian M, Rudenko A, Ye J, Zeng J, Huang W, Covey DF, Zhang R#, Fu Z#, Lishko PV#. Bioactive lipid-mediated structural and functional regulation of the essential human potassium channel Kir7.1. Nat Commun (2026).
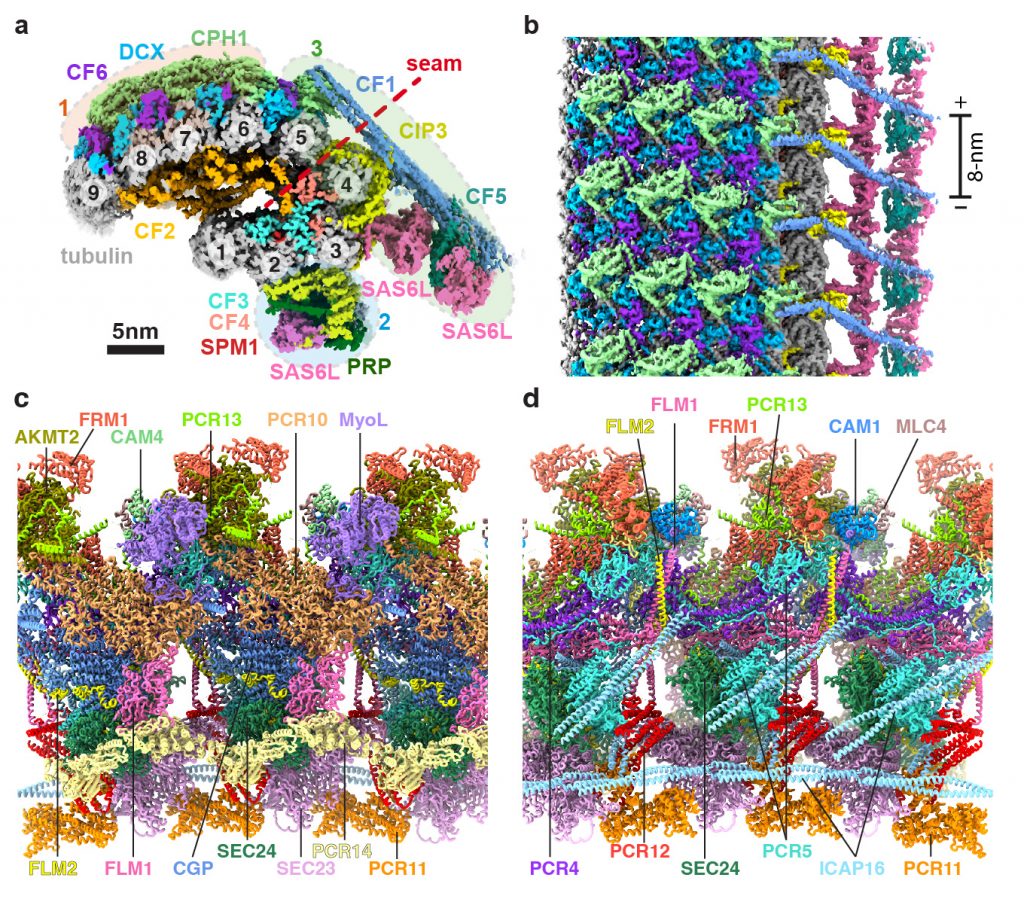
Zhang R, Sibley LD. Research Briefing: High-resolution cryo-EM meets parasitology in structural models of the conoid from Toxoplasma. Nat Struct Mol Biol (2026) 33, 5–6.
Hao L, Zhang R, Lohman TM. The Nuclease Domain of E. coli RecBCD Helicase Regulates DNA Binding and Base Pair Melting. J Mol Biol (2026) 438, 169571
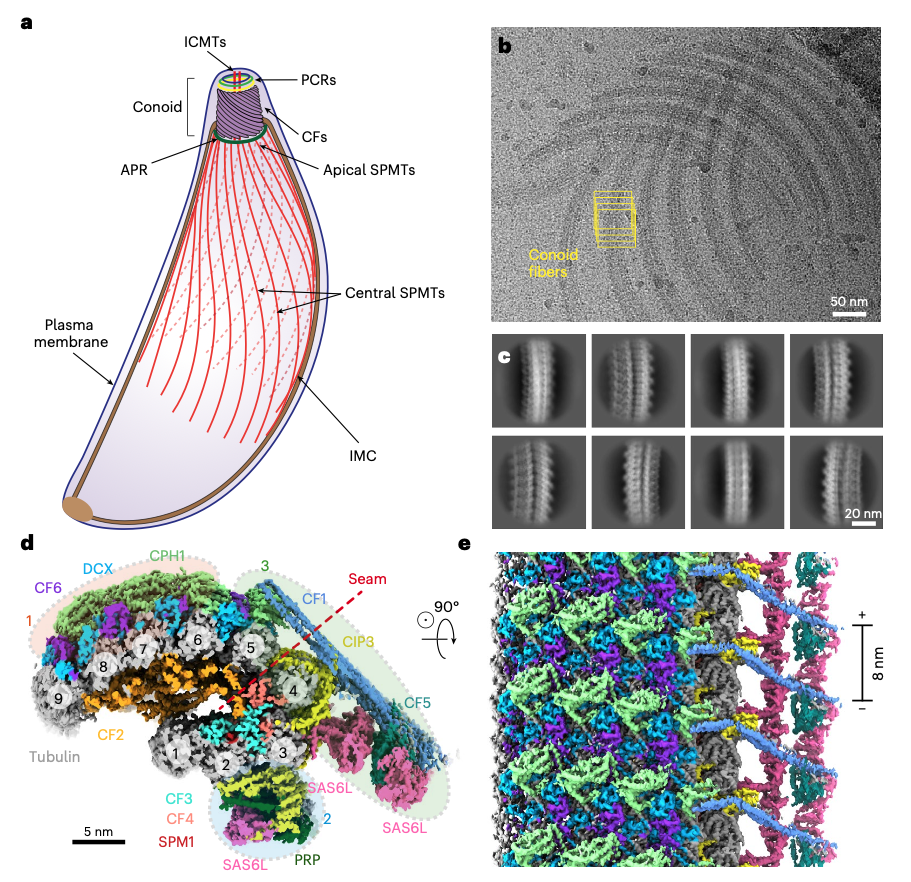
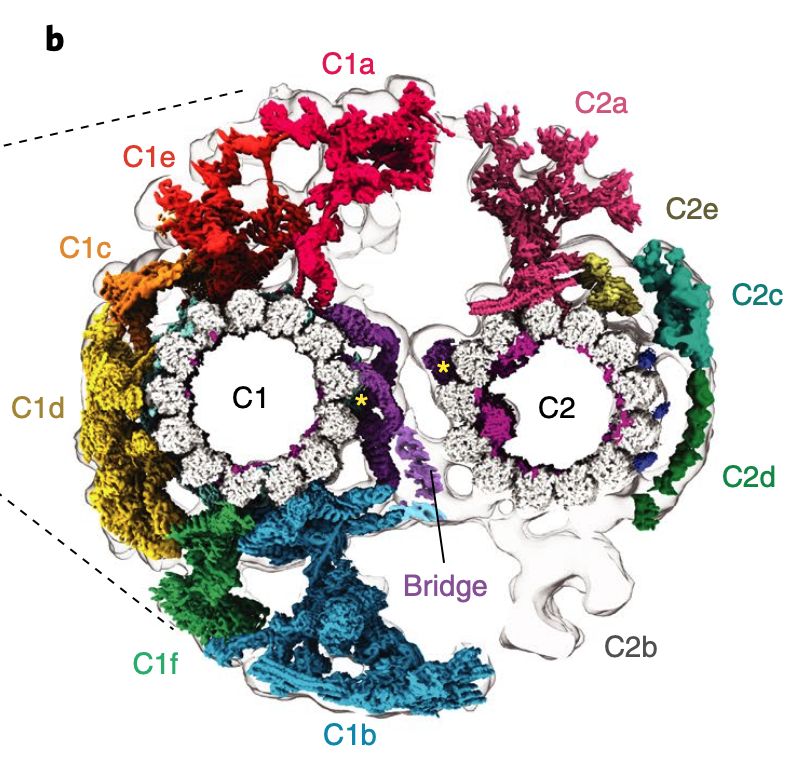
Zeng J*, Fu Y*, Qian P*, Huang W, Niu Q, Beatty WL, Brown A, Sibley LD#, Zhang R#. Atomic models of the Toxoplasma cell invasion machinery. Nat Struct Mol Biol (2026) 33, 157-170.
2025
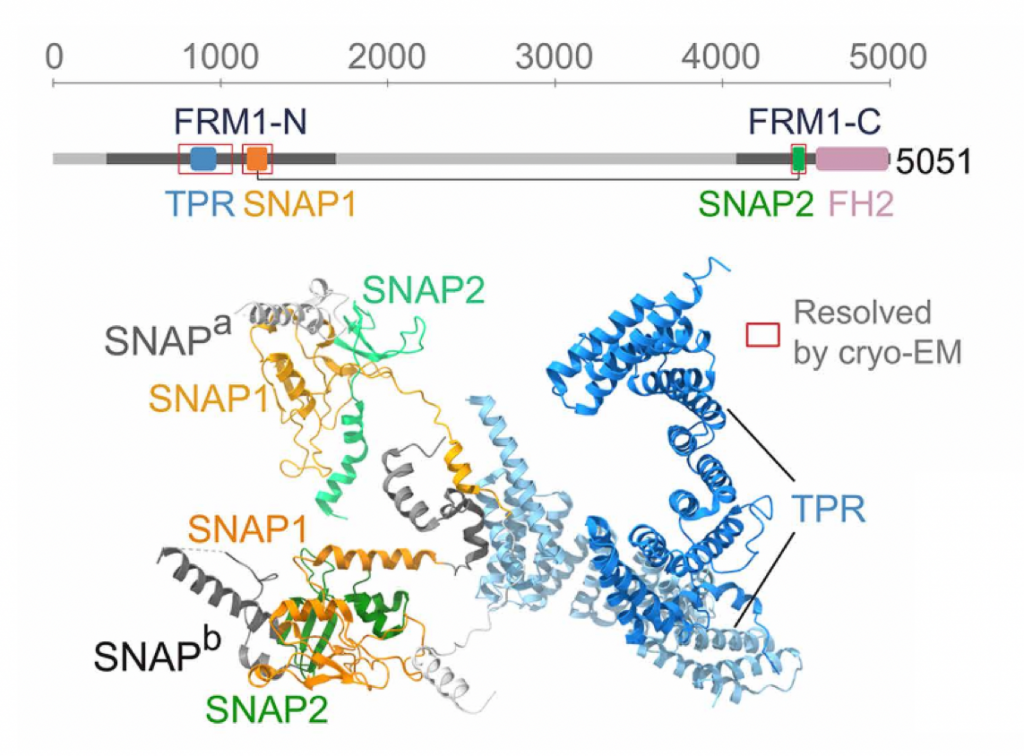
Qian P, Fu Y, Zeng J, Naldrett MJ, Zhang R#, Sibley LD#. Evolutionarily convergent mechanism of formin regulation coordinates actin polymerization in apicomplexan parasites. Sci Adv (2025) 11. eaea2136
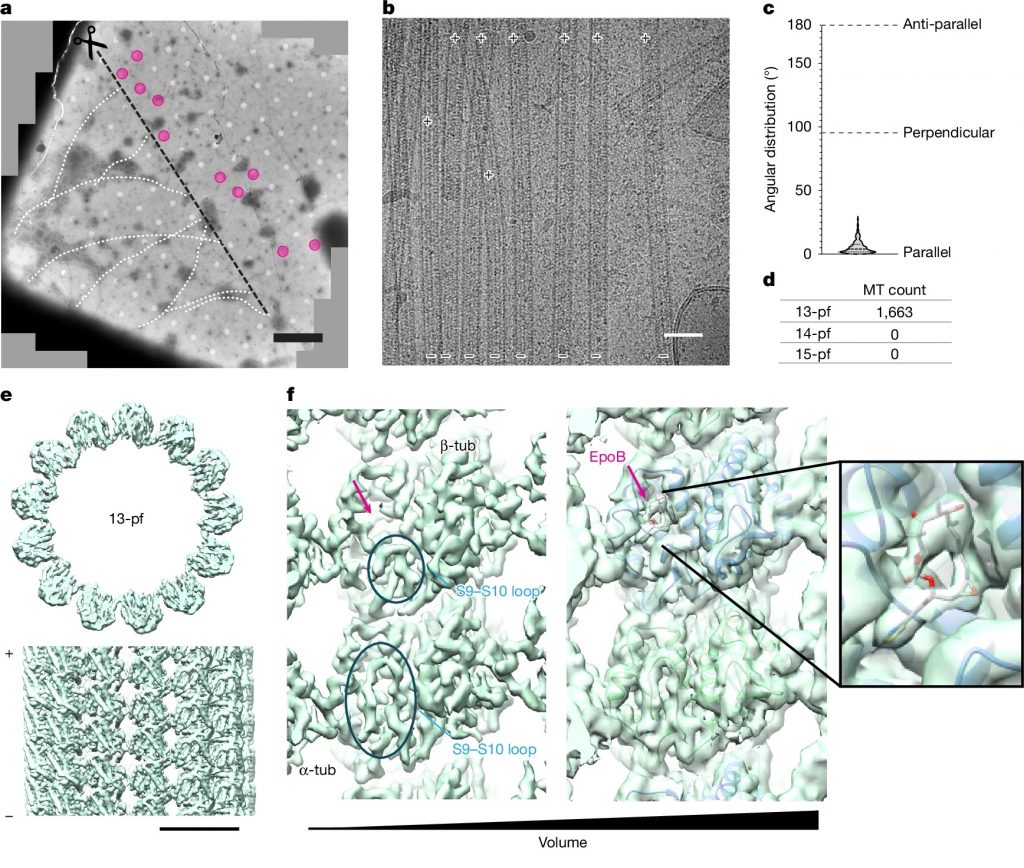
Bodakuntla S, Taira K, Yamada Y, Alvarez-Brecht P, Cada AK, Basnet N, Zhang R, Martinez-Sanchez A, Biertümpfel C, Mizuno N#. In situ structural mechanism of epothilone-B-induced CNS axon regeneration. Nature (2025) 648, 477–487.
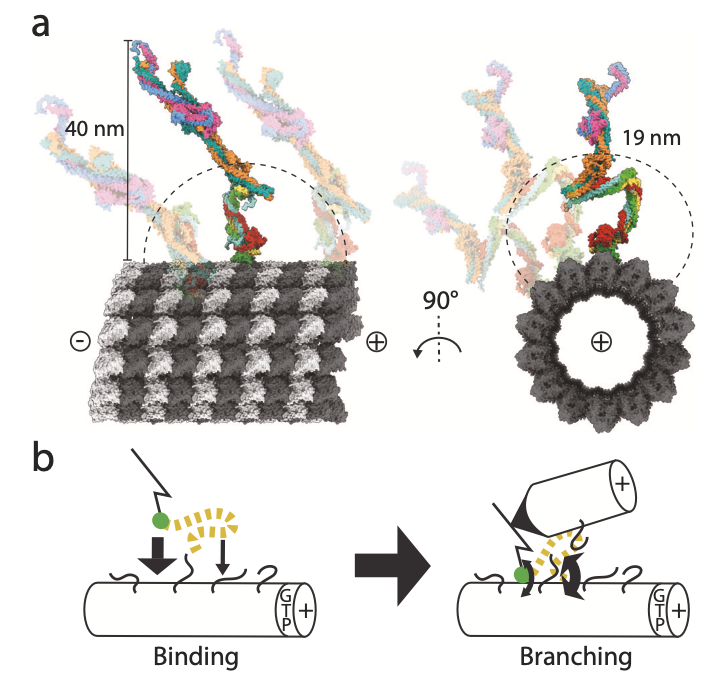
Travis SM, Kraus J, McManus CT, Golden K, Zhang R, Petry S#. How augmin establishes the angle of the microtubule branch site. Nat Commun (2025) 16, 9646.
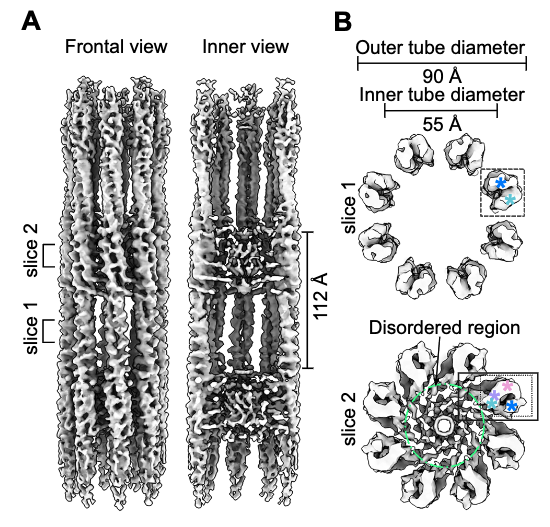
Agostini L, Pfister JA, Basnet N, Ding J, Zhang R, Biertümpfel C#, O’Connell KF#, Mizuno N#. Structural insights into SSNA1 self-assembly and its microtubule binding for centriole maintenance. Nat Commun (2025) 16, 7512.
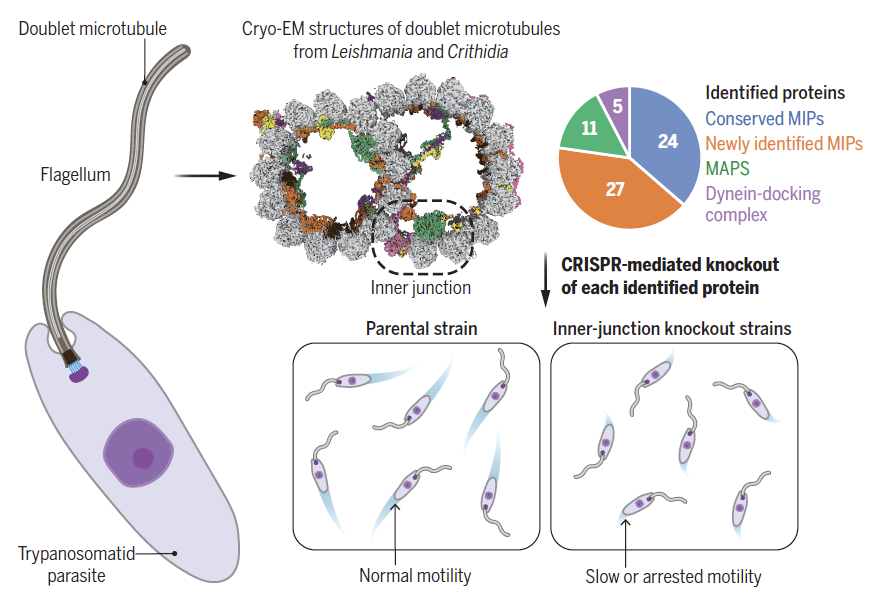
Doran MH*, Niu Q*, Zeng J, Beneke T, Smith J, Ren P, Coscia A, Höög JL, Meleppattu S, Lishko PV, Wheeler RJ, Gluenz E, Zhang R#, Brown A#. Evolutionary adaptations of doublet microtubules in trypanosomatid parasites. Science (2025) 387, eadr5507.
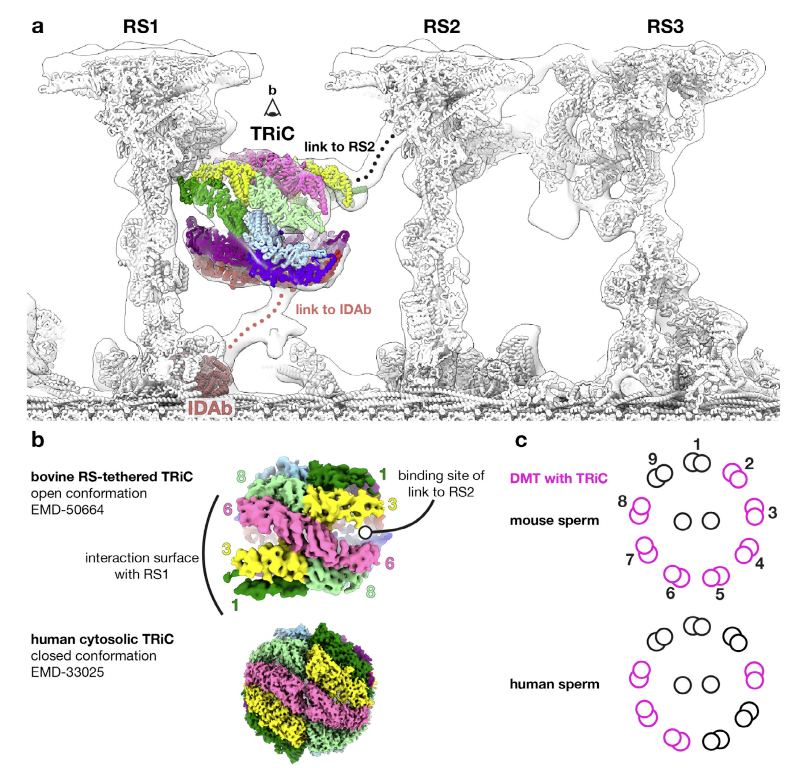
Brown A, Leung MR#, Zeev-Ben-Mordehai T, Zhang R. TRiC Is a Structural Component of Mammalian Sperm Axonemes. Cytoskeleton (2025).

Brody SL#, Pan J, Huang T, Xu J, Xu H, Koenitizer JR, Brennan SK, Nanjundappa R, Saba TG, Rumman N, Berical A, Hawkins FJ, Wang X, Zhang R, Mahjoub MR, Horani A, Dutcher SK#. Undocking of an extensive ciliary network induces proteostasis and cell fate switching resulting in severe primary ciliary dyskinesia. Sci Transl Med (2025) 17, eadp5173.
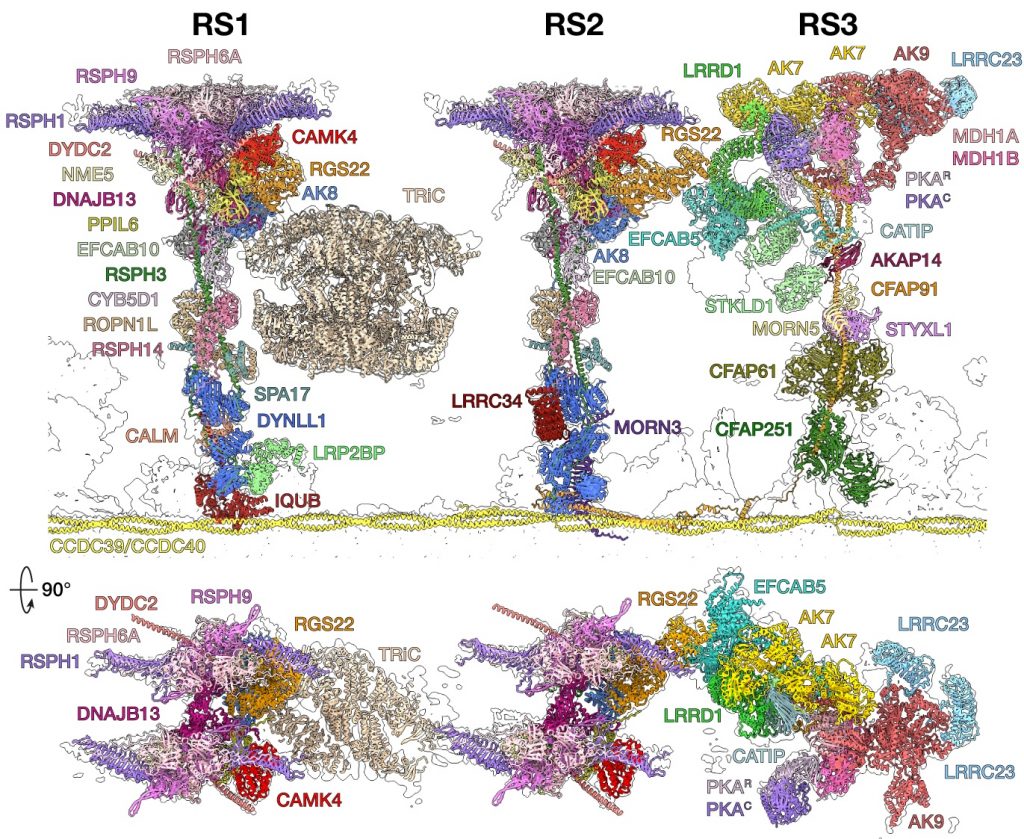
Leung MR*, Sun C*, Zeng J*, Anderson JR*, Niu Q, Huang W, Noteborn WEM, Brown A#, Zeev-Ben-Mordehai T#, Zhang R#. Structural diversity of axonemes across mammalian motile cilia. Nature (2025) 637, 1170–1177.
2024
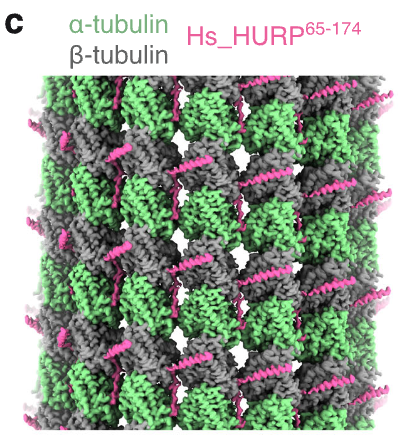
Valdez V, Ma M, Gouveia B, Zhang R#, Petry S#. HURP facilitates spindle assembly by stabilizing microtubules and working synergistically with TPX2. Nat Commun (2024) 15, 9689.
Garg A, Jansen S, Greenberg L, Zhang R, Lavine KJ, Greenberg MJ. The ACTA1 R256H mutation associated with dilated cardiomyopathy disrupts actin structure and function, leading to cardiomyocyte hypocontractility. Proc Natl Acad Sci USA (2024) 121(46): e2405020121.
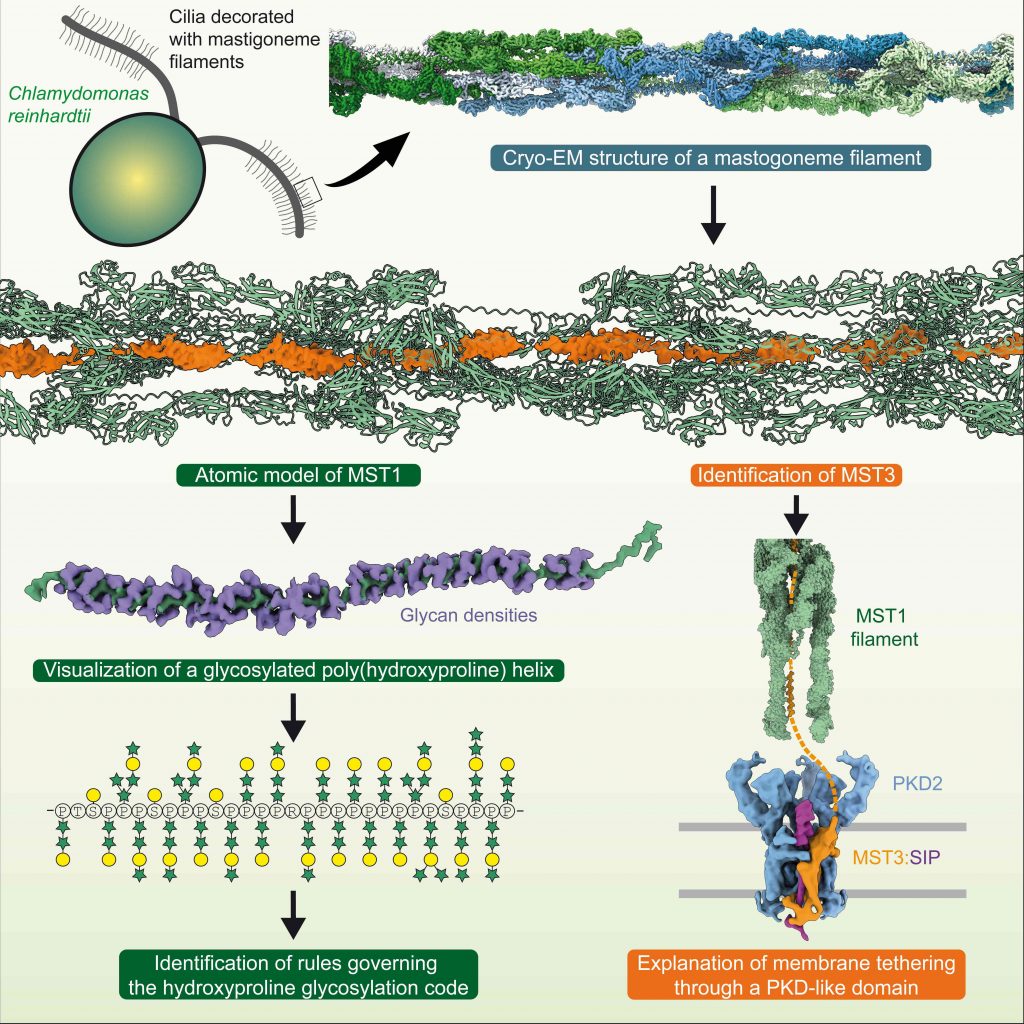
Dai J*, Ma M*, Niu Q, Eisert RJ, Wang X, Das P, Lechtreck KF, Dutcher SK, Zhang R#, Brown A#. Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices. Cell (2024) 187, 1907–21.
Jamali K#, Käll L, Zhang R, Brown A, Kimanius D#, Scheres SHW#. Automated model building and protein identification in cryo-EM maps. Nature (2024) 628, 450–457.
Strickland MR, Rau MJ, Summers B, Basore K, Wulf J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM. Apolipoprotein E secreted by astrocytes forms antiparallel dimers in discoidal lipoproteins. Neuron (2024) 112, 1100-1109.e5.
2023
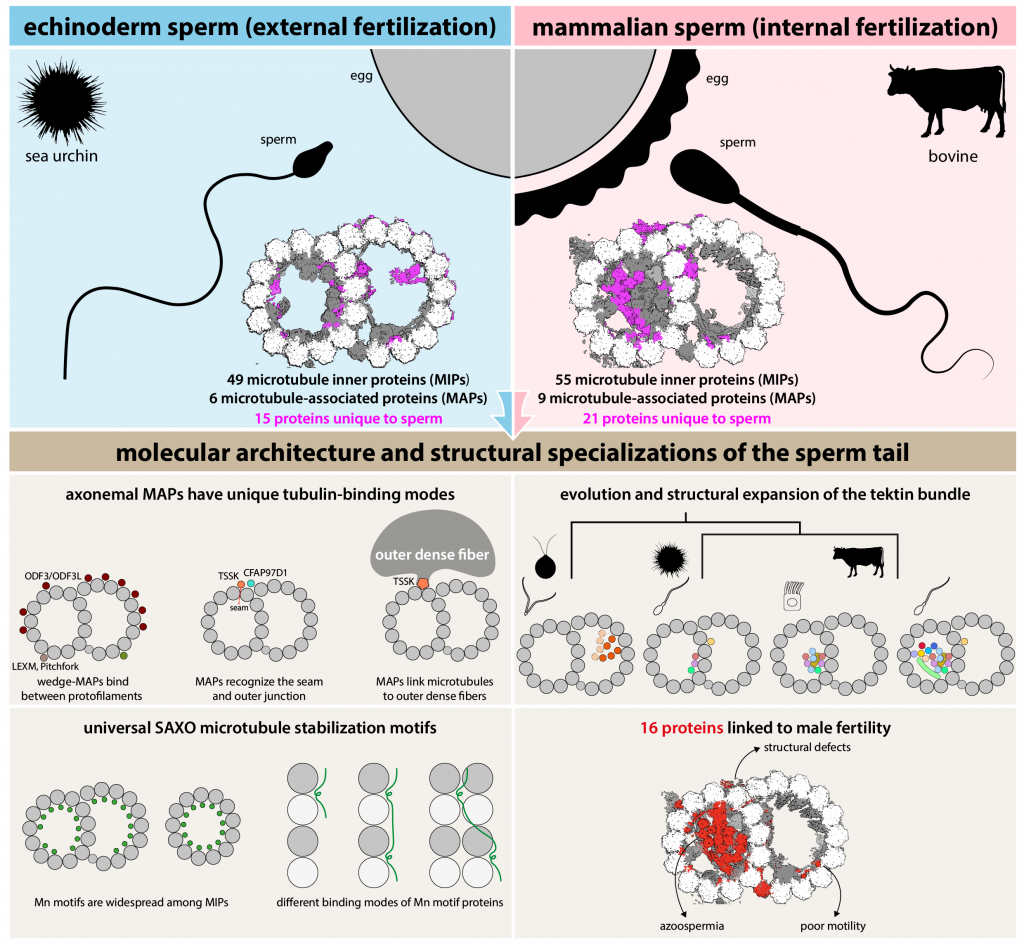
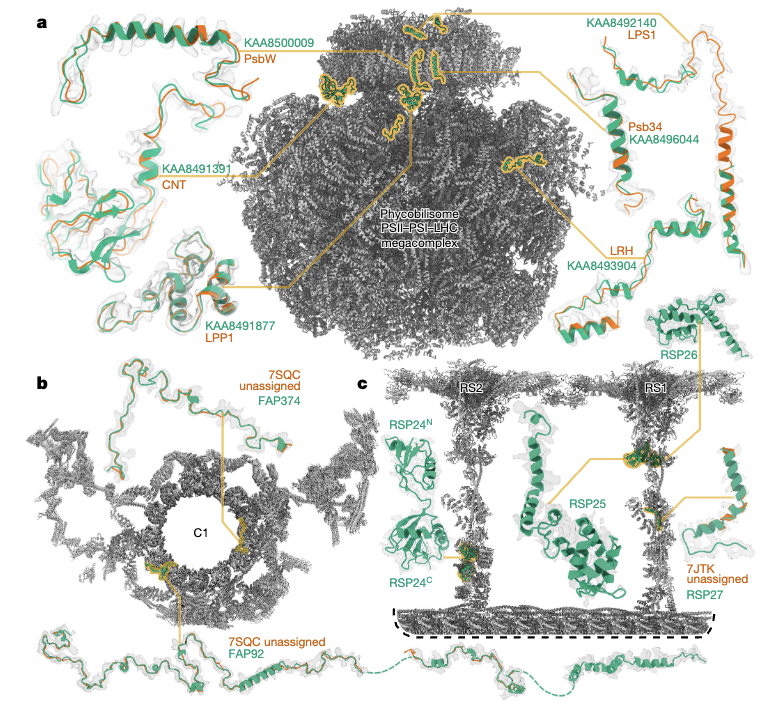
Leung MR*, Zeng J*, Wang X, Roelofs MC, Huang W, Chiozzi RZ, Hevler JF, Heck AJR, Dutcher SK, Brown A, Zhang R#, Zeev-Ben-Mordehai T#. Structural specializations of the sperm tail. Cell (2023) 186, 2880–2896.
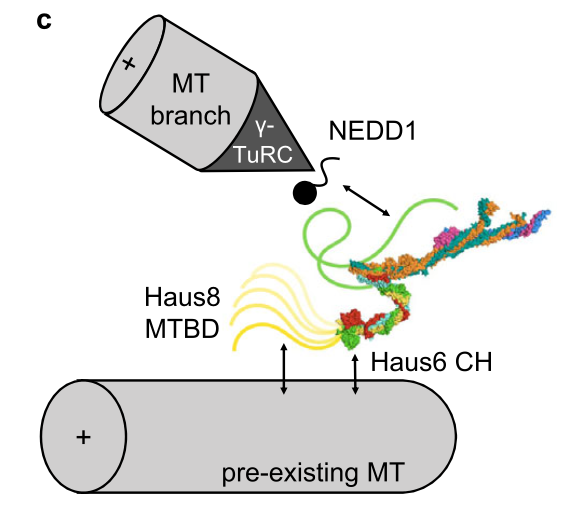
Travis SM*, Mahon BP*, Huang W, Ma M, Rale MJ, Kraus J, Taylor DJ, Zhang R#, Petry S#. Integrated model of the vertebrate augmin complex. Nat Commun (2023) 14, 2072.
2022
Gui M, Wang X, Dutcher SK, Brown A#, Zhang R#. Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Nat Struct Mol Biol (2022) 29, 483–492.
Hao L, Zhang R, Lohman TM. Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting. J Mol Biol (2022) 433 (18), 167147.
2021
Song X*, Yang F*, Liu X*, Xia P*, Yin W, Wang Z, Wang Y, Yuan X, Dou Z, Jiang K, Ma M, Hu B, Zhang R, Xu C, Zhang Z, Ruan K, Tian R, Li L, Liu T, Hill DL, Zang J#, Liu X#, Li J#, Cheng J#, Yao X#. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat Chem Biol (2021) 17, 1314–1323.
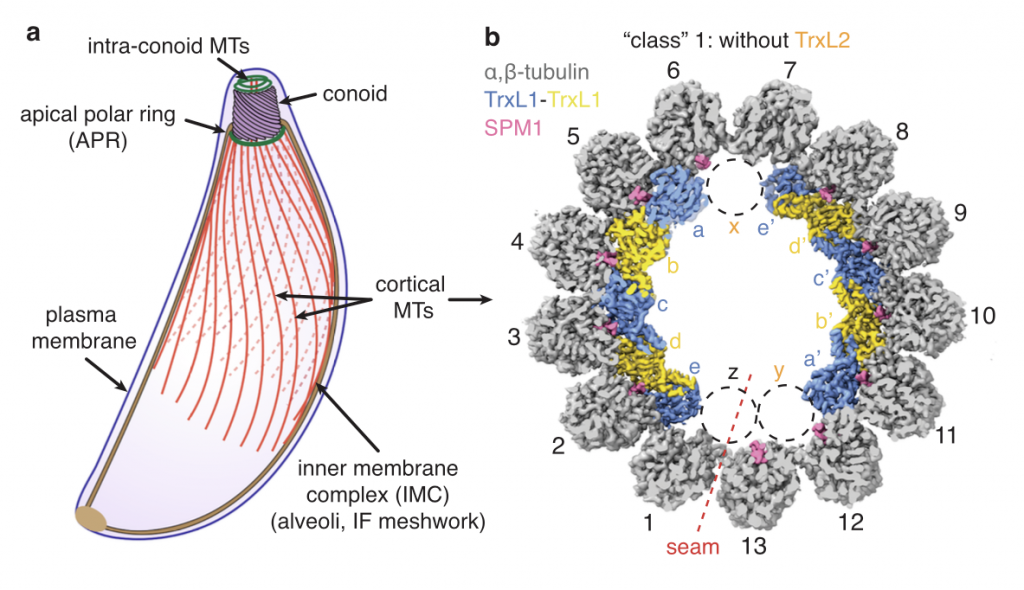
Wang X*, Fu Y*, Beatty WL, Ma M, Brown A, Sibley LD#, Zhang R#. Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins. Nat Commun (2021) 12, 4076–81.
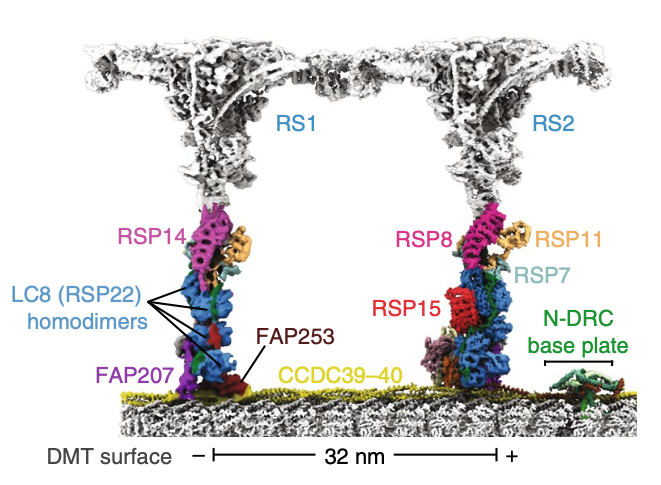
Gui M, Ma M, Sze-Tu E, Wang X, Koh F, Zhong ED, Berger B, Davis JH, Dutcher SK, Zhang R#, Brown A#. Structures of radial spokes and associated complexes important for ciliary motility. Nat Struct Mol Biol (2021) 28, 29–37.
2020
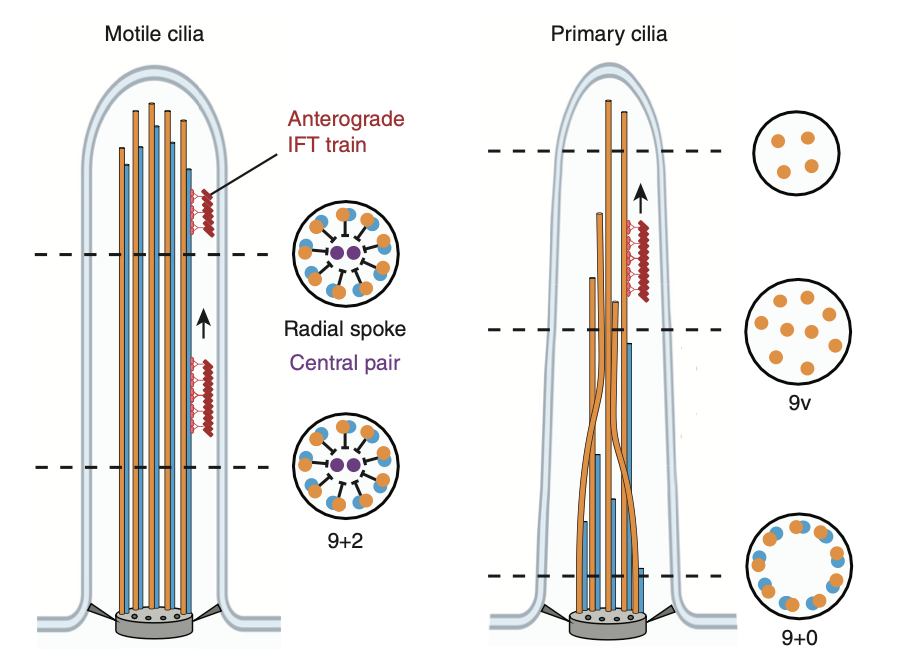
Brown A, Zhang R. Primary Cilia: A Closer Look at the Antenna of Cells. Curr Biol (2020) 30, R1494–R1496.
2019
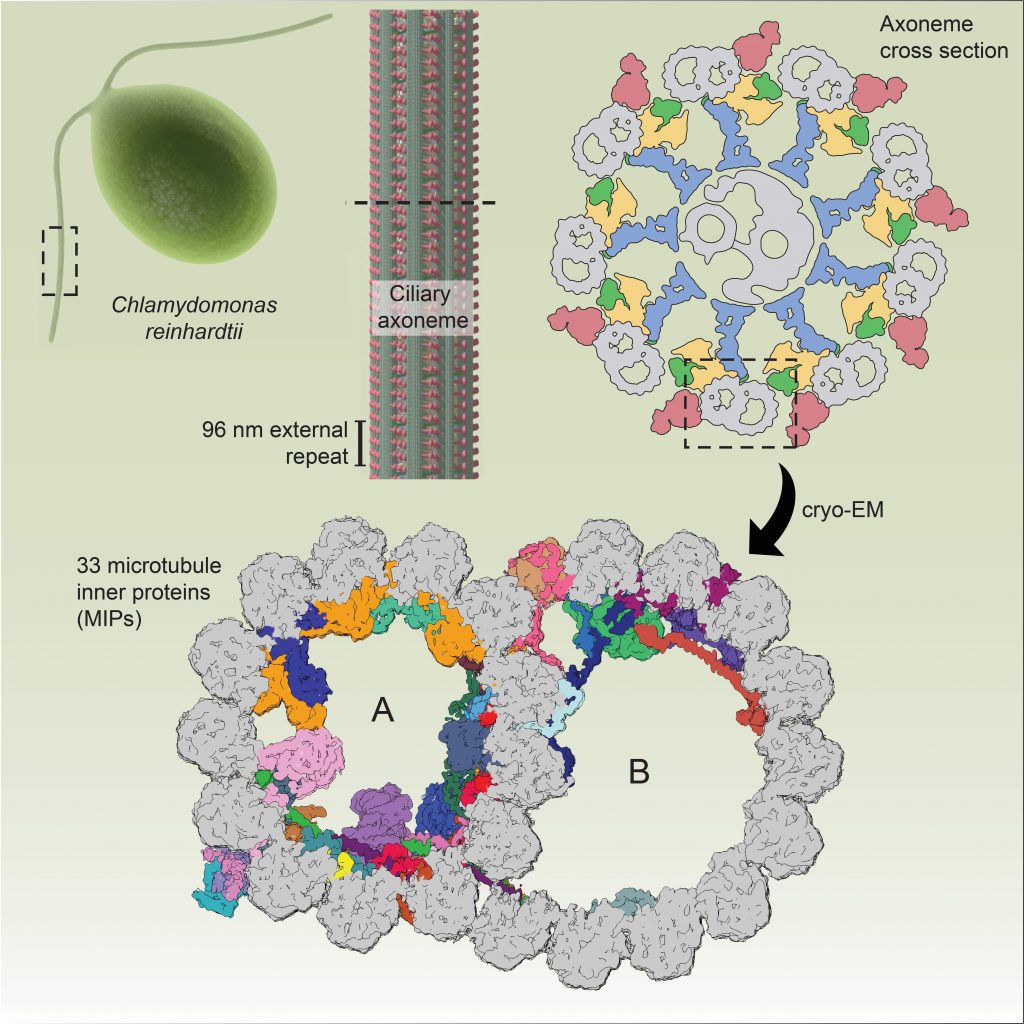
Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A#, Zhang R#. Structure of the decorated ciliary doublet microtubule. Cell (2019) 179, 909-12.
2018
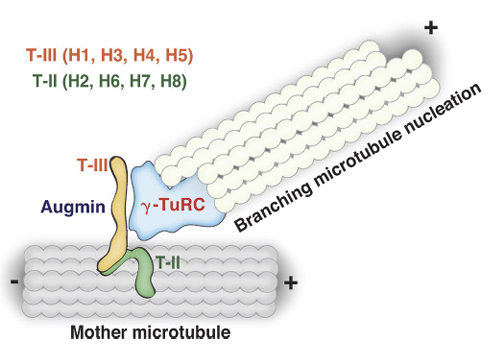
Song JG, King MR, Zhang R, Kadzik RS, Thawani A, Petry S. Mechanism of how augmin directly targets the γ-tubulin ring complex to microtubules. J Cell Biol (2018) 217 (7): 2417-2428.
Eva Nogales Lab @ UCB/LBNL
LaFrance BJ, Roostalu J, Henkin G, Greber BJ, Zhang R, Normanno D, McCollum CO, Surrey T#, Nogales E#. Structural transitions in the GTP cap visualized by cryo-electron microscopy of catalytically inactive microtubules. Proc Natl Acad Sci USA (2022) 119 (2), e2114994119.
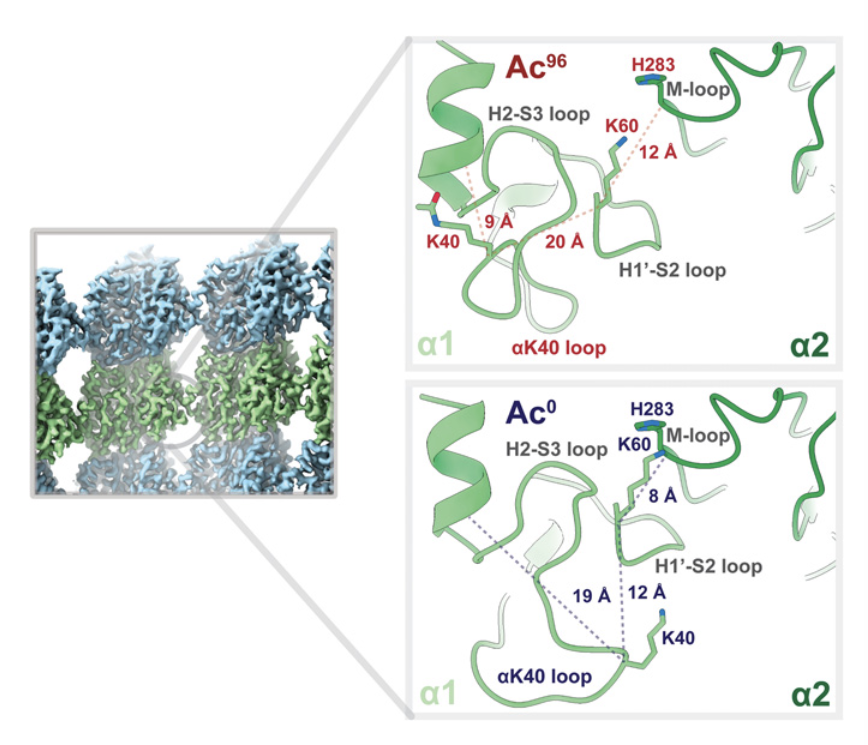
Eshun-Wilson L, Zhang R, Portran D, Nachury MV, Toso DB, Löhr T, Vendruscolo M, Bonomi M, Fraser JS#, Nogales E#. Effects of α-tubulin acetylation on microtubule structure and stability. Proc Natl Acad Sci USA (2019) 116 (21): 10366-10371.
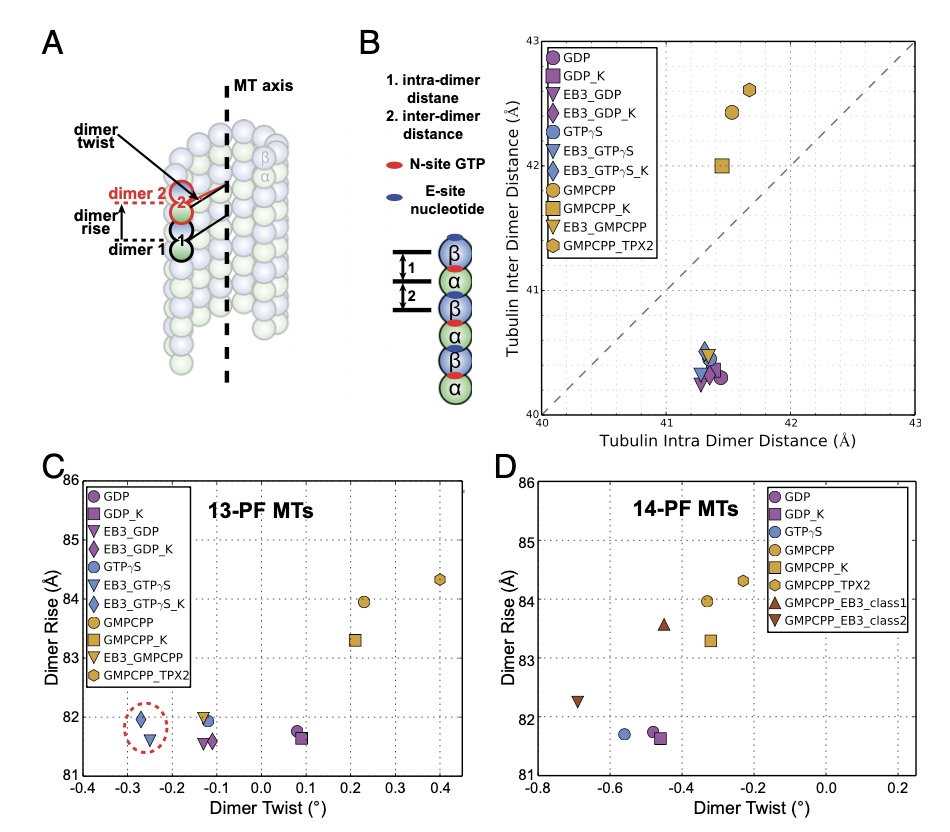
Zhang R#, LaFrance B, Nogales E#. Separating the effects of nucleotide and EB binding on microtubule structure. Proc Natl Acad Sci USA (2018) 115 (27): E6191-E6200.
Howes SC*, Geyer EA*, LaFrance B, Zhang R, Kellogg EH, Westermann S, Rice LM, Nogales E#. Structural and functional differences between porcine brain and budding yeast microtubules. Cell Cycle (2018) 17(3): 278-287.
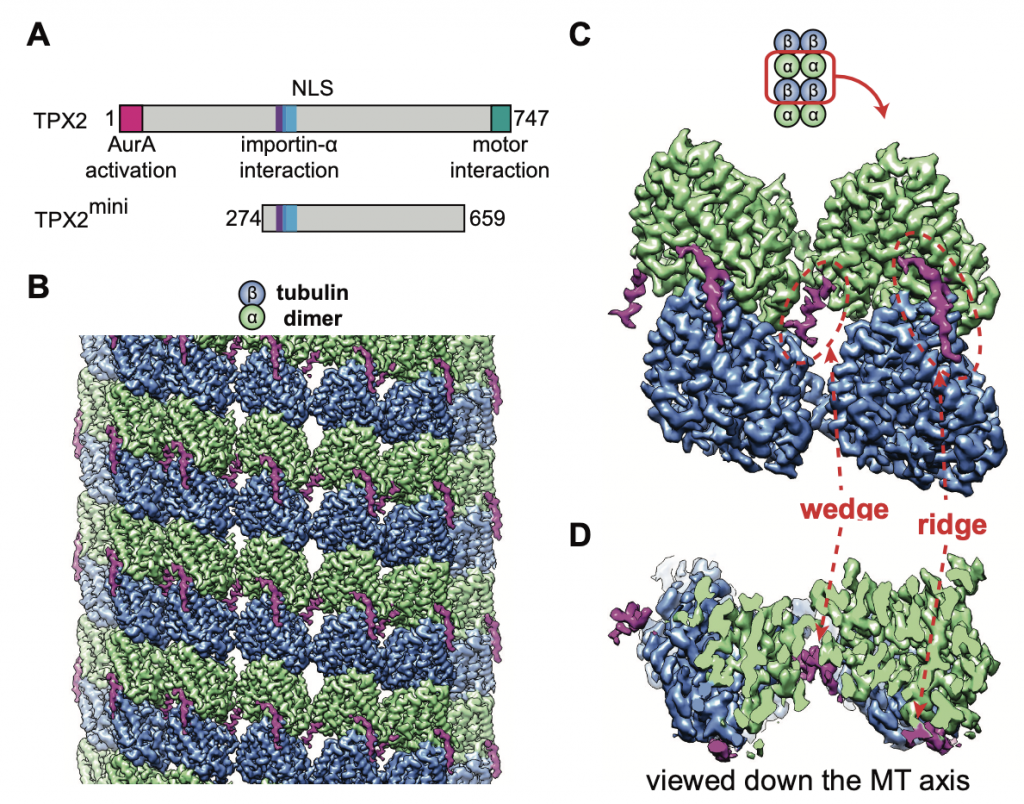
Zhang R*, Roostalu J*, Surrey T#, Nogales E#. Structural insight into TPX2-stimulated microtubule assembly. Elife (2017) 6: e30959.
Howes SC, Geyer EA, LaFrance B, Zhang R, Kellogg EH, Westermann S, Rice LM, Nogales E#. Structural differences between yeast and mammalian microtubules revealed by cryo-EM. J Cell Biol (2017) 216(9): 2669-2677.
Liu T, Dai A, Cao Y, Zhang R, Dong MQ, Wang HW. Structural insights into WHAMM’s interaction with microtubules by cryo-EM. J Mol Biol (2017) 429(9): 1352-1363.
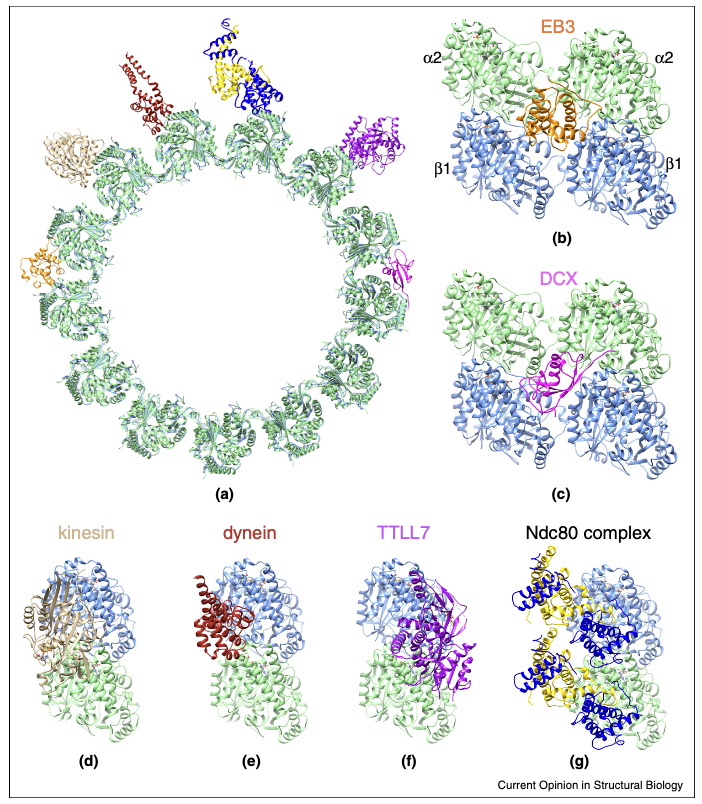
Nogales E, Zhang R. Visualizing Microtubule Structure and Interactions with Associated Proteins. Curr Opin Struct Biol (2016) 37: 90-96.
Feng R, Sang Q, Kuang Y, Sun X, Yan Z, Zhang S, Shi J, Tian G, Luchniak A, Fukuda Y, Li B, Yu M, Chen J, Xu Y, Guo L, Qu R, Wang X, Sun Z, Liu M, Shi H, Wang H, Feng Y, Shao R, Chai R, Li Q, Xing Q, Zhang R, Nogales E, Jin L, He L, Gupta ML, Cowan NJ, Wang L. Mutations in TUBB8 and Human Oocyte Meiotic Arrest. N Engl J Med (2016) 374: 223-232.
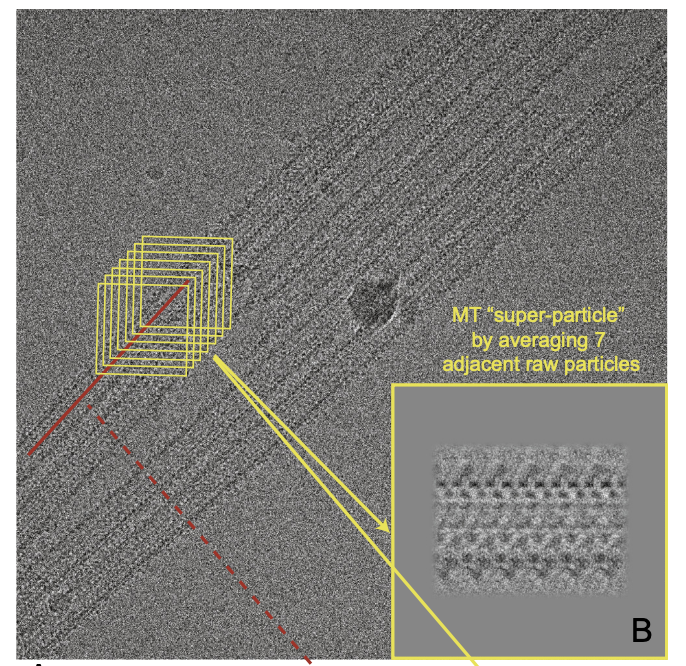
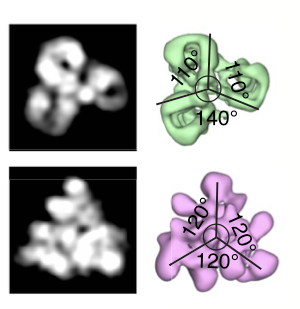
Zhang R#, Nogales E#. A New Protocol to Accurately Determine Microtubule Lattice Seam Location. J Struct Biol (2015) 192(2): 245-254.
Shen QT*, Ren X*, Zhang R*, Lee IH, Hurley JH. HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science (2015) 350(6259): aac5137.
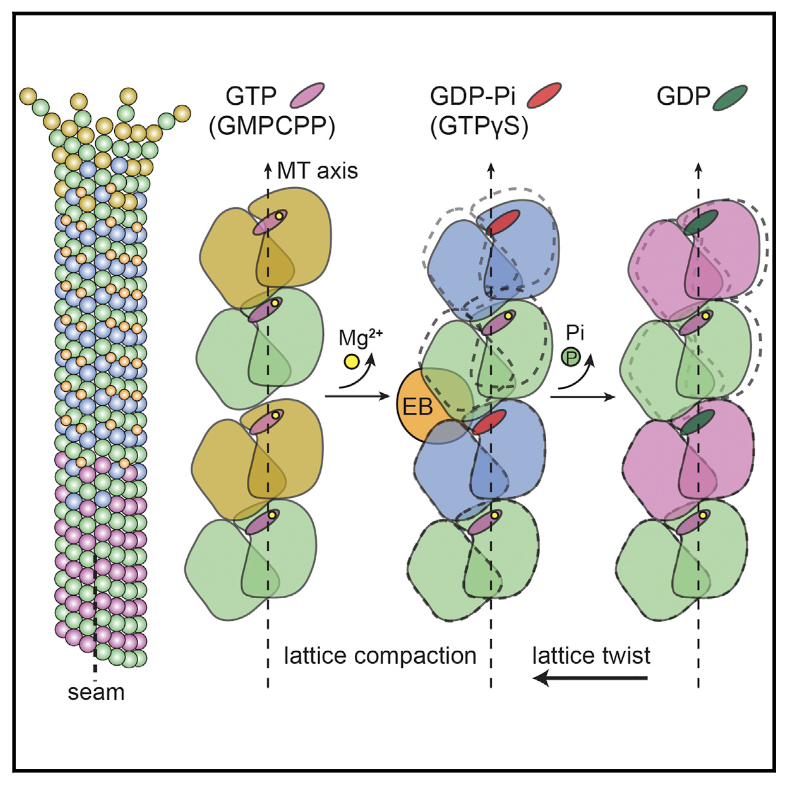
Zhang R, Alushin GM, Brown A, Nogales E#. Mechanistic Origin of Microtubule Dynamic Instability and its Modulation by EB Proteins. Cell (2015) 162, 849-859.
Alushin GM*, Lander GC*, Kellogg EH*, Zhang R, Baker D, Nogales E#. High resolution microtubule structures reveal the structural transitions in α/β tubulin upon GTP hydrolysis. Cell (2014) 157(5): 1117-1129.
Wah Chiu Lab @ BCM
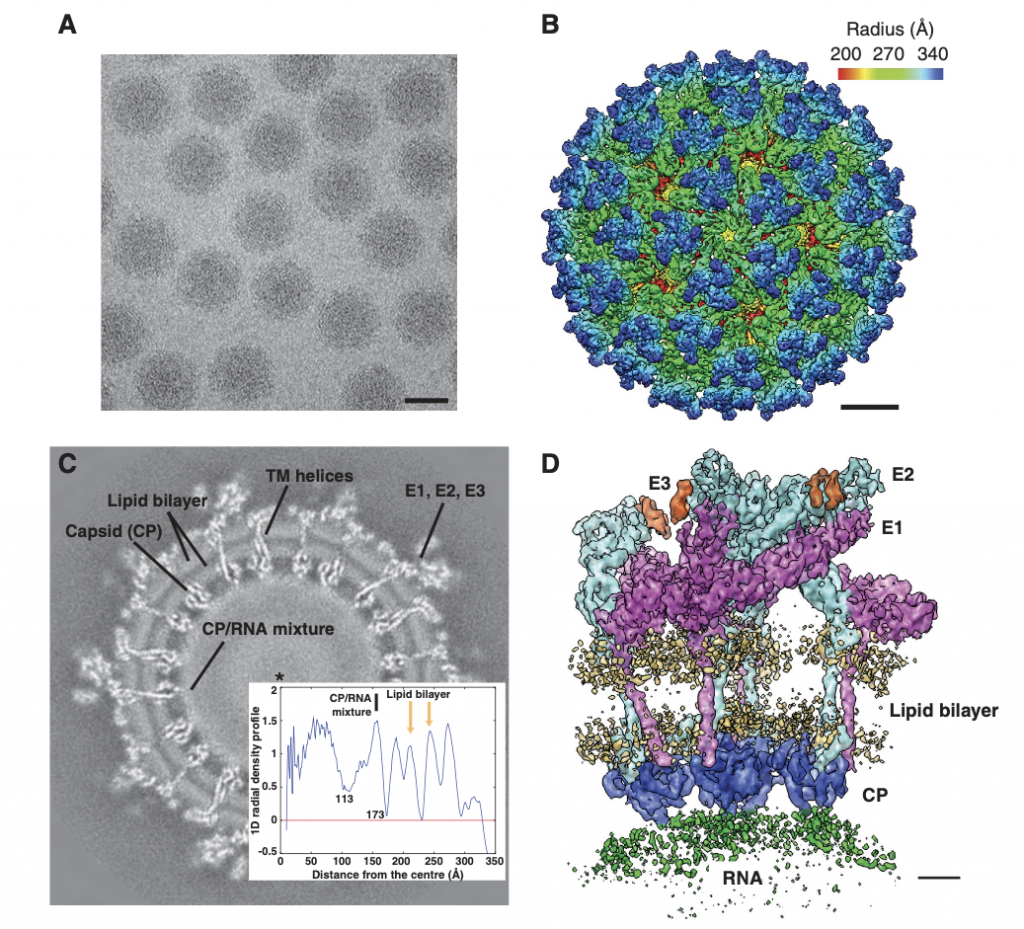
Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J (2011) 30(18): 3854-3863.
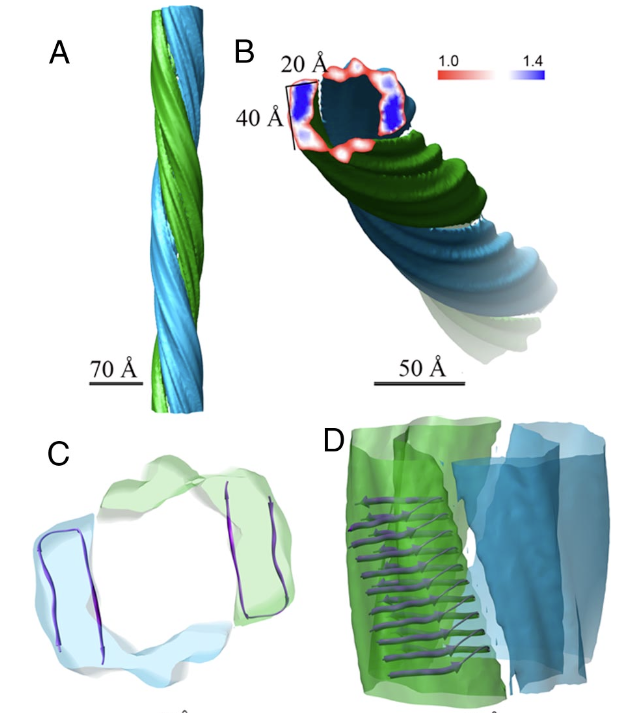
Zhang R, Hu X, Khant H, Ludtke SJ, Chiu W, Schmid MF, Frieden C#, Lee JM. Interprotofilament interactions between Alzheimer’s Aβ1-42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci USA (2009) 106: 4653-4658.
Parmenter CD, Cane MC, Zhang R, Stoilova-McPhie S. Cryo-electron microscopy of coagulation Factor VIII bound to lipid nanotubes. Biochem Biophys Res Commun (2008) 366(2): 288-93.